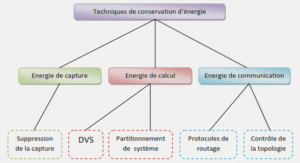
Sécheresse, irrigation et salinisation des sols
Materials and methods
Plant material
Seedlings were provided by the INRA-CIRAD station of Corsica, France. Diploid and autotetraploid seedlings of Willow leaf mandarin (Citrus deliciosa, SRA133) and trifoliate orange (Poncirus trifoliata (L.) cv. Pomeroy, SRA 1074) as well as their allotetraploid somatic hybrid FLHORAG1 (Ollitrault et al., 2000) were grown for 6 months in greenhouse. Identification of autotetraploid plants among diploids and diploid genetic conformity checking (nucellar status of the plants) were performed according to Mouhaya et al. (submitted for publication) using the same SSRs markers.
Twelve diploid and tetraploid plants of each genotype were then selected and transplanted on the same substrate in 3 L pots and grown in greenhouse. Plants were irrigated twice a week with half diluted nutritive solution [fertilizer 28-14-14 (ref 205), Fertile, France]. At one year, plants were pruned in order to homogenize the plant size of diploids and tetraploids.
Salt stress experiment
For each genotype, 5 two-years-old plants were selected for each condition. Salt stress was applied by watering plants daily at the same hour, either with 0.5L water supplemented with half diluted nutritive solution for control plants, or with 0.5L water supplemented with half diluted nutritive solution and 50 mM of NaCl for 2 weeks, then 100 mM for 2 weeks, then 200 mM for 2 weeks, and finally 400 mM for the last 2 weeks. By watering plants daily, pots were at the field capacity and the water potential was expected to be mostly dependent of the salt solution that was applied.
Experiments were performed during the summer in greenhouse in the following conditions
photoperiod (16/8), temperature (30 to 35°C) and relative humidity (60 to 85%).
Leaf water content, leaf thickness and maximum quantum yield of PSII analysis Leaf water content was monitored along the salt stress experiment at the same hour of the day: five leaves per genotype were harvested and fresh and dry weights were measured. Prior to be dried, leaf thickness of each genotype was measured by using a micrometer (Mitutoyo, IP65, Japan). Dry leaves were used for mineral analysis. Maximum quantum yield of PSII, (Fm- F0)/ Fm] was monitored along the same period by using a fluorescence meter (Hansatech Ltd, Kings Lynn,. UK). Maximum quantum yield of PSII was measured on three leaves of each genotype.
Mineral analysis: sodium and chloride content
Leaf samplings were performed along the stress experiment and root samplings were harvested at the end of the stress, for chloride and sodium content analysis. Those fractions were oven-dried at 60°C for one week, weighed, crushed in hammer-mill and stored at room temperature. Sodium and chloride analyses were carried out on dried leaves and roots according to Saleh et al. (2008).
For leaves, sodium and chloride contents were expressed in µg.mm-3 (taking account of leaf area and thickness) and in mg.g-1 of dry material for roots. ICP assays were performed at the US49, CIRAD-PERSYST in Montpellier, France.
Gene expression: Real time RT-PCR analysis
Samplings for gene expression analysis were regularly performed (7-days, 21-d, 35-d, 49-d, 56- d). Each sampling was a mix of leaves from four trees. Samplings were kept at minus 80°C. Total RNA were extracted from leaves collected after seven weeks (49-d) of the salt treatment according to Chomczynski and Sacchi (1987). Triplicate assays were performed on two independently collected leaves. Total RNA were purified using RNeasy Mini Kit (Qiagen, Leusden, The Netherlands) including an RNase free-DNase treatment using RNase free-DNase set (Qiagen, Leusden, The Netherlands). The total RNA concentration was measured at 260 nm and checked with RiboGreen RNA Quantitation Reagent and kit (Molecular Probes, Eugene, Oregon, USA) using FLUOstar Galaxy fluorometer (BMG Labtechnologies, Offenburg, Germany). Retrotranscription and Real time RT-PCR were performed together using the LightCycler 2.0 instrument (Roche, Diagnostics GmbH, Germany). Standard curves were done for all studied genes. To obtain standard curves, real time PCR was performed on dilutions of cDNA obtained by retrotranscription of 100 ng of a given RNA sample. 2.5 µL of DNase-treated total RNA (20ng/µL) were used in 10µL reactions containing 2µL of vial1 PCR mix (LightCycler FastStart DNA MasterPLUS SYBR Green I), 0.25µM gene-specific primers (Table 3), 2.5 units of SuperScript II Reverse Transcriptase (Applied Biosystems), 1 unit of RNase Inhibitor (Applied Biosystems). PCRs were conducted by the following steps: a first step of retro-transcription at 48°C for 30min followed by a temperature program strating with 10min at 95°C then 45 cycles consisting of 2s at 95°C, 10s at primer annealing temperature and 15s at 72°C, and finally the melting temperature 42s at 95°C. Internal control (a reproducible point in the gene-specific standard curve) and negative control (without RNA) were included for each PCR.
The primers that were used in real time RT-PCR (Table 3) were defined using the software Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) to select primer pairs with minimal number of potential primer dimers and primer hairpins as possible.
Statistical analysis
Data are expressed by the mean value ± SE. We used SIGMASTAT from SPSS (Chicago; www.spss.com_software_science) to analyze the data. The t test and ANOVA test were used to detect differences between the genotypes and the growing conditions at the usual probability level of P= 0.05.
Results
Tetraploid obtaining and genetic constitution analysis and plant growth Using flow cytometry, tetraploids were screened among plantlets from mother trees of the San Guiliano germplasm among the same set of plantlets than in Mouhaya et al. (submitted for publication). Removal of zygotic plants was as well performed according to Mouhaya et al. (submitted for publication). Only nucellar plants were kept for salt stress investigations. At one year, tetraploid plants were smaller than diploids. Plants were pruned in order to homogenise them. 10 two-years-old homogeneous plants of each genotype were selected for salt stress experiment.
|
Table des matières
AVANT-PROPOS
INTRODUCTION BIBLIOGRAPHIQUE
I. Sécheresse, irrigation et salinisation des sols
1.Problématique du manque d’eau dans les régions sèches
2.La pratique de l’irrigation et ses conséquences sur la qualité des sols
3.La salinisation des sols et la sensibilité des espèces cultivées
II. Les agrumes
1.Origine et taxonomie des agrumes
2.La diversité des agrumes et ses limites d’exploitation
3.La diversité des agrumes et la variabilité des résistances aux facteurs biotiques et
abiotiques
4.Le greffage des agrumes : moyen d’adaptation aux facteurs environnementaux
5.Accumulation des ions Na+ et Cl- chez les agrumes greffés : exemples d’association porte-greffe / greffon
6.Création de nouveaux porte-greffe cumulant des tolérances aux facteurs biotiques et
abiotiques
6.1. Création par hybridation sexuée
6.2. Création par hybridation somatique
III. Déroulement de la réponse au stress salin (NaCl) chez les plantes
IV. Déterminants physiologiques de la tolérance au stress salin chez les plantes : état de connaissances chez les agrumes
1.La croissance
2.La régulation stomatique et des échanges gazeux
3.La photosynthèse
V. Déterminants moléculaires de la tolérance au stress salin chez les plantes : état de connaissance chez les agrumes
1.La perception du stress salin
2.La transduction du signal du stress salin
2.1. Les cations Ca2+
2.2. L’hormone ABA (Acide ABscissique)
2.3. Les Espèces actives de l’oxygène ‘ROS’
3.Les cascades de signalisation
3.1. La voie Salt Overly Sensitive ‘SOS’
3.2. La voie Mitogen-Activated Protein Kinase ‘MAPK’
3.3. Les phosphatases
4.Les facteurs de transcription
4.1. Les protéines à doigt de zinc
4.2. Les protéines à doigt de zinc de type WRKY
4.3. Les protéines Myb
5.Activation de l’expression de gènes pour l’adaptation au stress salin
5.1. Osmoprotection
5.1.1. La glycine-bétaïne
5.1.2. La proline
5.2. Les protéines chaperonnes : Late Embryogenesis Abundant ‘LEA’
5.3. La régulation de l’homéostasie ionique
5.3.1. Transport de Na+
5.3.2. Transport de Cl-
5.4. L’homéostasie hydrique : les aquaporines
5.5. La détoxication antioxydante
5.5.1. L’acide ascorbique et l’Ascorbate peroxydase
5.5.2. Le glutathion et la glutahion réductase (GR)
5.5.3. La superoxyde dismutase (SOD)
5.5.4. La catalase (CAT)
VI. La polyploïdie et son impact chez les plantes
1.La polyploïdie
2.La polyploïdie chez les plantes
3.Impacts de la polyploïdie chez les plantes
3.1. Impact phénotypique
3.2. Impact génomique
4.Impact de la polyploïdie chez les agrumes
5.Amélioration de la tolérance des agrumes aux stress via l’hybridation somatique
VII. Présentation des travaux de thèse
MATERIEL ET METHODES
I. Matériel végétal
II. Conditions de croissance et application du stress salin
III. Méthodologie
1.Caractérisation génétique des agrumes polyploïdes
1.1. Recherche des individus autotétraploïdes : cytométrie en flux
1.2. Recherche et élimination des individus zygotiques
2.Caractérisation de l’allotétraploïde FLHORAG1
2.1. Comptage chromosomique
2.2. Héritabilité des génomes nucléaire et cytoplamiques chez le FLHORAG1
2.2.1. Extraction et quantification de l’ADN total
2.2.2. Amplification par réaction de polymérisation en chaîne PCR
2.2.3. Electrophorèses sur gel de polyacrylamide dénaturants et non dénaturants
3.Anatomie foliaire chez les agrumes polyploïdes
3.1. Mesure de la surface stomatique
3.2. Mesure de l’épaisseur foliaire
3.3. Mesure de la croissance
3.4. Dosage du taux de chlorophylle
4.Déterminants physiologiques du stress salin chez les agrumes polyploïdes
4.1. Mesure de l’efficacité du photosystème II
4.2. Dosage minéral : concentrations foliaires et racinaires en Na+ et en Cl-
5.Etude transcriptomique de la réponse au stress salin chez les agrumes polyploïdes
5.1. Extraction des ARN totaux
5.2. Analyse transcriptomique par la technique de cDNA-AFLP (Amplified Fragment Length Polymorphism)
5.2.1. Principe de la technique cDNA-AFLP
5.2.2. Technique
5.2.2.1. Isolement des ARNm et synthèse des ADNc double brin
5.2.2.2. La réaction AFLP
5.2.2.3. Electrophorèse sur gel de polyacrylamide dénaturant
5.2.2.4. Isolement et séquençage des TDFs (Transcript Derived Fragments)
5.2.2.5. Analyse fonctionnelle des TDFs
5.3. Analyse transcriptomique par PCR quantitative : RT-PCR en temps réel
5.3.1. Principe
5.3.2. Technique
RESULTATS
I. Résultats sous forme d’articles
1.Article 1
2.Article 2
II. Résultats complémentaires
1.Etude des déterminants physiologiques de la tolérance à la salinité chez différentes associations porte-greffe / greffon
1.1. Epaisseur des feuilles et contenu en eau
1.2. Fluorescence chlorophylienne de feuilles, [(Fm-F0)/Fm]
1.3. Contenus racinaires et foliaires en ions chlorures
2.Etude des déterminants moléculaires de la tolérance au stress salin chez les différentes associations porte-greffe / greffon
DISCUSSION GENERALE
I. Effet de l’auto- et l’allopolyploïdisation chez les porte-greffe d’agrumes en condition de non stress
II. Effet de l’auto- et l’allopolyploïdisation chez les porte-greffe d’agrumes en condition de stress salin
1.Le porte-greffe Poncirus trifoliata
2.La variété mandarinier commun
3.Le FLHORAG1
III. Réponse des variétés greffés sur les porte-greffe autotétraploïdes de Poncirus et sur l’allotétraploïde FLHORAG1
CONCLUSION ET PERSEPECTIVES
REFERENCES BIBLIOGRAPHIQUES
ANNEXES
![]() Télécharger le rapport complet
Télécharger le rapport complet