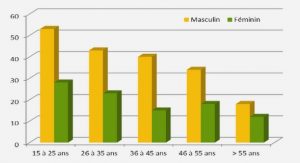
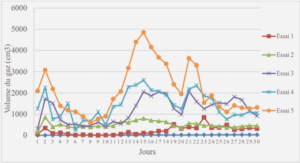
Phylogenetic analysis of the AP2 domain from AP2/ERF marker genes
Identification of 35 marker genes of somatic embryogenesis capacities during somatic embryogenesis expression/induction
The calli transferred to a somatic embryogenesis induction medium also strongly expressed a large number of the genes studied for virtually all the families and groups, except for the ERF-V group (Figure 4 A). As previously, most of the members of groups I (9 genes), VII (12 genes) and VIII (10 genes) of the ERF family had a high transcript level. The differential expression between calli of the different lines was significant for 35 genes (Figure 4 B). Of those genes, 15 had higher transcript accumulation in the regenerant line than in the NE and E lines: 1 ethylene biosynthesis gene (HbSAMS), 11 genes of the ERF family (HbERF-IIIb2, HbERF-IIIc1, HbERF-IIIe3, HbERF-IVa1,HbERF-IVa2, HbERF-IVa3, HbERF-Vb2, HbERF-VI-L1, HbERF-VIIIa7, HbERF-IXb2, HbERF-IXc4), 2 genes of the AP2 family (HbAP2-3, HbAP2-7) and 1 gene of the RAV family (HbRAV-3). Transcripts of the HbERF-VIIIa3 gene were accumulated in the calli of the embryogenic line compared to the other two lines. Lastly, 19 genes comprising 14 ERFs (HbERF-IIIa1, HbERF-Vb3, HbERF-VI3, HbERF-VI4, HbERF-VI-L6, HbERF-VIIa3, HbERF-VIIa4, HbERF-VIIa13, HbERF-VIIa17, HbERF-VIIIa4, HbERF-VIIIa13, HbERF-IXa2, HbERF-IXa3, HbERF-IXa4) and 5 AP2s (HbAP2-4, HbAP2-6, HbAP2-8, HbAP2-11, HbAP2-19) had a lower transcript level in the non-embryogenic line than in the others.
Sixteen genes were differentially expressed in normal and abnormal somatic embryos
The normal and abnormal somatic embryos produced by the regenerant line revealed a differential expression profile solely for certain members of the ERF family (Figure 5). The gene transcripts belonging to each of the families and groups were highly accumulated, except for the RAV family (Figure 5 A). Most of the members of the ERF-I, ERF-VII and ERF-VIII groups were highly expressed in the two types of embryos. The transcript abundance was lower in the normal embryos than in the abnormal embryos for 12 genes belonging to the ERF family (HbERF-Ia1, HbERF-Ib1, HbERF-Ib3, HbERF-Ib6, HbERF-Ib7, HbERF-Ib8, HbERF-IIb5, HbERF-IVa2, HbERF-VIIIa11, HbERF-IXb4, HbERF-IXb8, HbERF-Xb1) (Figure 5 B). Conversely, the transcripts of 4 genes were accumulated in the normal embryos (HbERF-VI1, HbERF-VIIa1, HbERF-VIIa3, HbERF-VIIa4).
Evolution of transcript abundance during the process of somatic embryogenesis and conversion into plantlets
An overall analysis of the somatic embryogenesis process for the regenerant line using proliferating callus up to the conversion of normal embryos into plantlets showed that the expression of numerous genes was highly modulated (Figure 6). The transcripts of numerous ethylene biosynthesis and transduction genes, and some ERFs from groups I, VII and VIII and of the AP2 family, were highly accumulated (Figure 6 A). Thirty-six genes from all the families or groups revealed differential accumulation during the embryogenesis process, except for the ERFs from group X (Figure 6 B). Several gene expression profiles were observed. Eleven markers were activated right from the somatic embryogenesis induction phase (HbSAMS, HbACO1, HbEIN3, HbERF-IIIe2, HbERF-VIIa9, HbERF-VIIa23, HbERF-VIIIa3, HbERF-IXb2, HbAP2-5, HbAP2-11, HbAP2-13).
|
Table des matières
List of figures
List of tables
Liste des abréviations
Introduction générale
1 Hevea brasiliensis and natural rubber industry
2 Regulation of latex production in Hevea brasiliensis
3 Genetic improvement of rubber tree
4 Ethylene biosynthesis and signaling in plants
5 AP2/ERF superfamily
6 Towards the characterization of the ethylene biosynthesis and signalling in Hevea brasiliensis
References
CHAPITRE 1 Article intitulé “Some ethylene biosynthesis and AP2/ERF genes reveal a specific pattern of expression during somatic embryogenesis in Hevea brasiliensis”
Abstract
Background
Results
1 Morphogenetic potential of callus lines with different somatic embryogenesis capacities
2 Identification of 14 marker genes of somatic embryogenesis capacities during callus proliferation
3 Identification of 35 marker genes of somatic embryogenesis capacities during somatic embryogenesis expression/induction
4 Sixteen genes were differentially expressed in normal and abnormal somatic embryos
5 Evolution of transcript abundance during the process of somatic embryogenesis and conversion into plantlets
6 Identification of putative functions for somatic embryogenesis marker genes
Discussion
Conclusion
Methods
1 Plant material
2 Total RNA isolation
3 Primer design and analysis of transcript abundances by real-time RT-PCR
4 Statistical data analyses
5 Phylogenetic analysis of the AP2 domain from AP2/ERF marker genes
References
CHAPITRE 2 Article intituled “Hevea brasiliensis has a large Ethylene Response Factor group VII, which is highly expressed in latex cells”
Abstract
Background
Results
1 Functional annotation and classification of a comprehensive Hevea transcriptome
2 Relative transcript abundance in various tissues
3 Identification of putative functions for HbERF genes
4 Gene structure and in silico promoter analysis of some HbERF-VIIs
5 Functionality of HbERF-VIIs
Discussion
Material and methods
1 Plant material
2 Total RNA isolation
3 Sequencing technique and contig assembly
4 Primer design and analysis of transcript abundances by real-time RT-PCR
5 Statistical data analyses
6 Phylogenetic analysis of the AP2 domain from ERF marker genes
7 Cis-acting element analysis and determination of conserved motifs
8 Subcellular localization and transcriptional activity tests by transient expression in a single cell system
References
Discussion générale
1 Enrichment of the RNAseq database for the improvement of the AP2/ERF sequences
2 Identification of AP2/ERF putatively involved in important function in Hevea
3 Functional analysis of AP2/ERF putatively involved in important function
4 Use of expression marker genes
5 Conclusion
References
![]() Télécharger le rapport complet
Télécharger le rapport complet