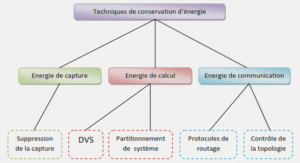
NON-RIGID 2D/3D REGISTRATION OF CORONARY ARTERY MODELS WITH LIVE FLUOROSCOPY FOR GUIDANCE OF CARDIAC INTERVENTIONS
Medical imaging segmentation
A rich literature on image segmentation has emerged in the past 25 years, framed around a variety of mathematical paradigms. Among the methods that have had the most notable influence on the field, we note the snakes method (Kass et al., 1988), based on differential geometry and a variational framework; the level-set and fast marching methods (Osher and Sethian, 1988; Sethian, 1996), which take advantage of implicit representations of continuous functions; the graph cut methods (Greig et al., 1989; Roy and Cox, 1998; Boykov et al., 2001; Kolmogorov, 2004), involving a type of combinatorial optimization; the random walkers (Grady, 2006) and power watershed methods (Couprie et al., 2011), rooted in graph and probability theory; and methods based on probabilistic and non parametric clustering, such as the mean shift algorithm (Comaniciu and Meer, 2002), the MPM-MAP algorithm (Marroquín et al., 2002), and a regression of a Gaussian Mixture Model (Greenspan et al., 2006). The more fundamental work of Mumford and Shah (1989) contributed to our understanding of the segmentation problem.
Vessel detection filters
Vessel detection and segmentation is a challenging task for a variety of reasons: the structures are small and complex, their observed shape can vary, pathologies may be present that affect their geometry, and contrast is limited in medical imaging, to name a few. At the same time, automatic vessel detection has many potential clinical applications. For example, it can help diagnose vascular disease by highlighting regions with arteriogenesis or stenosis, and facilitate surgical mapping and guidance. Because of the complexity of the structures of interest, many dedicated algorithms have been proposed in the literature. The vessel detection filter methods constitute one type of approach that is of interest. Theses methods use local features to compute a vesselness measure, which represent the level to which a pixel belongs to the vessel class. The application of a vessel detection filter on a 3D scan highlights vessel-like structures and discards the background, providing enhanced visualization. Vessel detection filters are also a basic building block in many complex medical image processing applications (van Bemmel et al., 2003; Sundar et al., 2006; Ruijters et al., 2009; Schneider and Sundar, 2010).
Types of 2D/3D registration methods
It is possible to classify the various 2D/3D registration methods that have been described in the literature essentially by inspecting two of their most important characteristics: 1) what is the basis for comparison, and 2) what is the dimensionality of the comparison domain 2. The basis for comparison refers to the abstractions that are actually compared during the registration process. We consider two classes of basis for comparison for both the 2D and 3D modalities: image bases and model bases. An image basis involves simply the input images, either the 2D fluoroscopies or the 3D volume, with or without pre processing with some low level filter, such as a gradient or vesselness filter. In contrast, a model basis is composed of some features of interest. For the 2D modality, this basis could be, for example, the thresholding of a vesselness filter response or the result of a structure identification process. For the 3D modality, the basis could consist of a 3D segmentation of the structures of interest, a centerline representation or a more complex model. The dimensionality of the comparison domain refers to the dimension of the similarity measurement space. The comparison domain is either 2D or 3D. Bidimensional comparison domains are associated with projective methods. The key aspect of these methods is that the 3D representation is projected onto the 2D imaging planes as part of the comparison process. The 3D to 2D projection can be simulated X-ray fluoroscopies, called digitally reconstructed radiographs, or a maximum intensity projection. The methodology is reversed when a 3D comparison domain is chosen
Image-to-image methods
Image-to-image projective 2D/3D registration methods are the most frequently encountered methods in the literature (Markelj et al., 2012). They involve projecting the volumetric information onto the 2D imaging plane. In general, the physical properties of the imaged tissues, as well as the characteristics of the imaging source, are modeled and encoded into the projection algorithm. In this case, the projected volume results in a synthetic X-ray fluoroscopic image called a Digitally Reconstructed Radigraph (DRR), which is very similar to a real 2D fluoroscopic image (Hipwell et al., 2003). Other projection algorithms, such as maximum intensity projection, have been considered by some authors (Kerrien et al., 1999). In this category of registration methods, the two principal research directions have been the efficient creation of high-quality DRRs (Khamene et al., 2006), and the definition of the best performing comparison metrics (Liao et al., 2006; Birkfellner et al., 2009).A related registration technique is the image-to-image reconstruction-based 2D/3D registration method of Tomazevic et al. (2006), in which a synthetic volume is reconstructed using multiple 2D angiography images acquired with a 3D rotational X-ray device, and the filtered backprojection technique of Grass et al. (1999).
Model-to-image methods
The problem of the 2D/3D registration of a 3D model of the aorta and attached MAPCAs has been addressed by our team in Julien Couet and et al. (2012). In this research, DDRs were generated using a fast wobbled splatting technique (Birkfellner et al., 2005). A problem that generally occurs with this method is that a flies screen-lika pattern might appears on the DRRs because of the magnification induced by the simulated imaging system. We proposed to alleviate this problem by using a triangle convolution filter, which under certain circumstances is equivalent to using a 3D image upsampled by trilinear interpolation (Cosman, 2000). The minimization is driven by an improved hill climbing algorithm. This method resulted in a target registration error of less than 2.060±2.712mm on a difficult real-world MAPCAs dataset. The main drawback of this method is that the relatively high computational time required makes it more suitable for off-line applications.Registration methods based on the projection of full volumes or dense models on the 2D imaging plane always involve relatively high computational cost.
Model-to-model methods
Model-to-model 2D/3D registration methods are potentially much faster during the optimization step. Feldmar et al.describe a framework for the registration of points, lines, and surfaces in (Feldmar et al., 1994, 1995, 1997). The registration transformation is first approximated using geometrical analysis and is refined in a second step involving the Iterative Closest Point (ICP) algorithm. They present results obtained on a variety of datasets, including brain vasculature. Their method requires user interaction at the initialization level, which is a limitation for some applications, and they do not clearly describe how the features are extracted from the various images. More recently, Benameur et al. (2003) proposed a method of the registration of a deformable 3D mesh model of the vertebra with radiographic images. In this work, the features are automatically extracted in 2D using Canny’s edge detector. The registration parameters are computed using a gradient descent approach following an initial alignment step. An efficient model-to-model 2D/3D registration method for coronary artery centerline model alignment with x-ray images have been proposed by Sundar et al. (2006). This method is based on a Distance Transform (DT) of the 2D features on the image plane, a concept previously introduced by Paragios et al. (2003) in the context of 2D/2D registration. The main advantages of this method are its relatively large capture range and its computational efficiency.
|
Table des matières
INTRODUCTION
0.1 Clinical context
0.2 Problem statement
0.3 Outline of the thesis
CHAPTER 1 LITERATURE REVIEW
1.1 Medical imaging segmentation
1.1.1 The level-set method
1.1.2 Level-set segmentation of vascular structures
1.1.3 Brain tissue segmentation
1.2 Vessel detection filters
1.2.1 Analysis of the Hessian
1.2.2 Optimally oriented flux
1.2.3 Polar intensity profile
1.3 2D/3D registration of medical imaging
1.3.1 Types of 2D/3D registration methods
1.3.2 Image-to-image methods
1.3.3 Model-to-image methods
1.3.4 Model-to-model methods
CHAPTER 2 OBJECTIVES AND GENERAL METHODOLOGY
2.1 Objectives of the research
2.2 General methodology
2.2.1 Large structures segmentation using the level-set method
2.2.2 Vessel detection and segmentation
2.2.3 2D/3D registration for the guidance of surgical procedures
CHAPTER 3 UNSUPERVISED MRI SEGMENTATION OF BRAIN TISSUES USING A LOCAL LINEAR MODEL AND LEVEL SET
3.1 Introduction
3.1.1 State-of-the-art techniques and their limitations
3.1.2 The need for a more comprehensive model
3.1.3 Organization of this paper
3.2 A local linear level set approach to MRI segmentation
3.2.1 Formulation of the problem
3.2.2 Local linear region model
3.2.3 Segmentation using the level set method
3.2.4 Extension to a 4 phase model
3.2.5 Outliers rejection
3.2.6 Implementation issues
3.2.7 3D extension of the model
3.2.8 Computational cost
3.2.9 Initialization by Fuzzy C-means clustering
3.2.10 Overview of our new segmentation model
3.3 Experimental validation
3.3.1 Synthetic Data
3.3.2 Real Data
3.4 Discussion and conclusions
CHAPTER 4 3D VESSEL DETECTION FILTER VIA STRUCTURE-BALL ANALYSIS
4.1 Introduction
4.2 Methodology
4.2.1 The structure ball: a local structure model
4.2.2 Band-limited spherical harmonics representation
4.2.3 Contrast-invariant diffusivity index
4.2.4 Geometric descriptors
4.2.4.1 Ratio descriptors
4.2.4.2 Oriented bounding box
4.2.4.3 Fractional anisotropy
4.2.4.4 Flux
4.2.5 Vesselness measure
4.2.6 Multiscale integration
4.2.7 Parameter selection
4.3 Experimental results
4.3.1 Experiments on synthetic images
4.3.2 Experiments on clinical images
4.4 Discussion and conclusion
4.5 Acknowledgement
CHAPTER 5 NON-RIGID 2D/3D REGISTRATION OF CORONARY ARTERY MODELS WITH LIVE FLUOROSCOPY FOR GUIDANCE OF CARDIAC INTERVENTIONS
5.1 Introduction
5.2 Background Information
5.3 Translational, rigid, and affine alignment
5.3.1 Multi frame alignment
5.4 Non-rigid registration
5.4.1 Image energy
5.4.2 Internal energy
5.4.3 Energy minimization
5.4.4 Parameter selection
5.5 Experimental results
5.5.1 Simulations
5.5.1.1 Dependence on the initial solution
5.5.1.2 Robustness to image noise
5.5.1.3 Non-rigid deformation
5.5.2 Clinical data
5.5.3 Global alignment: evaluation of the performance of the optimizers
5.5.4 Comparison of the global alignment method with non-rigid registration
5.5.5 Global alignment in the multi frame scenario
5.5.6 Semiautomatic tracking of the right coronary artery
5.6 Multimedia Material
5.7 Discussion and conclusion
CHAPTER 6 DISCUSSIONS
6.1 A level-set method using local-linear region models for the segmentation of structures with spatially varying intensity
6.2 Brain tissues segmentation in MRI
6.3 Accurate local structure modeling and vessel detection using structure balls
6.4 2D/3D registration of centerline models with X-ray angiography using global transformation models
6.5 Non-rigid 2D/3D registration of centerline models with X-ray angiography
6.6 Improved surgical guidance using 2D/3D registration
6.7 Application for MAPCAs procedure
GENERAL CONCLUSION
![]() Télécharger le rapport complet
Télécharger le rapport complet