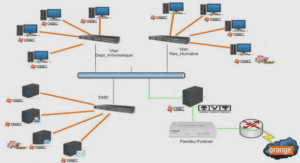
Methylation profiles of IL33 and CCL26 in bronchial epithelial cells are associated with asthma
Methylation profiles of IL33 and CCL26 in bronchial epithelial cells are associated with asthma
Avant-propos
Ce chapitre expose les principaux résultats du projet de recherche visant à documenter les patrons de méthylation et d’expression des gènes IL33, IL1RL1 et CCL26 dans des cellules épithéliales bronchiques dans un contexte d’asthme.
Les analyses ont permis de démontrer une différence significative du niveau de la méthylation des cytosines du promoteur des gènes IL33 et CCL26. La méthylation de l’ADN dans la région du promoteur du gène IL33 et trois polymorphismes ont aussi démontré une corrélation positive significative. Les résultats ont donc permis de confirmer la pertinence de l’étude de l’IL-33 et de sa voie biologique afin de mieux comprendre et éventuellement traiter la maladie.
Cet article est actuellement en révision et la soumission a été faite le 12 février 2018. La Dre Catherine Laprise a dirigé l’ensemble du projet et réalisé le devis de l’étude. Les biopsies bronchiques d’où ont été extraites les cellules épithéliales ont été réalisées à l’Institut Universitaire de Cardiologie et de Pneumologie de Québec (IUCPQ) par le Dr Michel Laviolette. Sophie Plante, de l’équipe du Dre Jamila Chakir de l’IUCPQ, s’est occupée de la mise en culture et de l’isolation des cellules épithéliales bronchiques. Sophie Plante et Valérie Gagné-Ouellet ont fait l’extraction de l’ADN et de l’ARNm ainsi que de la mesure de l’expression du gène IL1RL1. Valérie Gagné-Ouellet a également fait la conversion au bisulfite de sodium et le pyroséquençage du gène IL33. Par la suite, j’ai réalisé le pyroséquençage des deux autres gènes, IL1RL1 et CCL26, la transcription inverse, les PCR en temps réel pour la mesure de l’expression des gènes IL33 et CCL26, puis le séquençage de ces deux gènes. Marie-Chantale Larose, assistante de recherche du Dr Nicolas Flamant de l’IUCPQ, a contribué aux étapes pour le CCL26 en ayant mis au point les conditions pour la mesure d’expression. Suite à ces différentes manipulations, Valérie Gagné-Ouellet et moi avons rédigé l’article en collaboration. Catherine Laprise a dirigé les analyses statistiques, retravaillé le manuscrit. Tous les auteurs ont révisé le manuscrit et approuvé sa soumission. Catherine Laprise a finalement soumis l’article à la revue Epigenetics le 12 février 2018.
Résumé
L’épithélium des voies respiratoires joue un rôle important dans la réponse inflammatoire dans l’asthme. Les cellules épithéliales bronchiques relâchent des médiateurs pro-inflammatoires qui recrutent à leur tour des cellules inflammatoires telles que les éosinophiles. Malgré le fait que le rôle des cytokines relâchées par l’épithélium ait été largement décrit, les mécanismes régulant leur sécrétion demeurent toujours incertains. La méthylation de l’ADN, un mécanisme épigénétique, a précédemment été identifiée comme médiateur de la production de plusieurs cytokines de type auxiliaire 2 (TH2) dans les cellules sanguines. L’objectif de cette étude est de caractériser les variations de méthylation de l’ADN dans les gènes IL33, IL1RL1 et CCL26 dans des cellules épithéliales bronchiques d’individus asthmatiques et de voir les impacts fonctionnels sur la transcription de ces gènes. Le bis-pyroséquençage et le qRT-PCR ont été réalisés sur un échantillon de 10 patients asthmatiques et 10 témoins. Le séquençage des gènes a aussi été réalisé pour identifier des polymorphismes pouvant interagir avec les variations épigénétiques. Un niveau de méthylation de l’ADN plus faible a été détecté chez les asthmatiques pour les gènes IL33 et CCL26 (Δβ=15%, p<0.001 et Δβ=5%, p=0.036, respectivement). De plus, les individus porteurs d’un allèle mutant au sein d’un haplotype dans le promoteur d’IL33 (formé par chr9:6210100, rs928413 et rs7848215) avaient un niveau plus faible de méthylation (Δβ=6%, p=0.021 et Δβ=14%, p=0.038 pour le CpG 1 et la moyenne CpGs 2-3-4, respectivement), suggérant qu’une potentielle interaction entre ces SNP et le niveau de méthylation du promoteur d’IL33 pourrait mener à une action synergique augmentant le risque de développer de l’asthme. Le niveau d’expression des trois gènes était supérieur chez les asthmatiques, mais le seuil de significativité n’a pas été atteint. Par contre, une association entre le niveau d’expression de CCL26 et le décompte d’éosinophiles sanguins a été observée (r=0,772; p=0,009). Ces résultats démontrent l’importance d’investiguer à la fois les mécanismes génétiques et épigénétiques (par exemple les polymorphismes, l’expression génique et la méthylation de l’ADN) dans le but de mieux comprendre et de documenter la réponse immunitaire des cellules épithéliales bronchiques dans l’asthme.
Abstract
Airway epithelium plays a key role in the inflammatory response in asthma. Bronchial epithelial cells (BECs) release pro-inflammatory mediators, which recruit inflammatory cells such as eosinophils. Although the roles of epithelial cytokines have widely been described, the underlying mechanisms regulating their secretions remain unclear. The epigenetic mechanism of DNA methylation (DNA-me) has previously been identified to mediate the production of several type 2 helper cell (TH2) cytokines in blood cells, and this study aimed to characterize DNA-me variations in IL33, IL1RL1 and CCL26 in asthma and their functional impacts on transcriptional activity in BECs. We performed bis-pyrosequencing and qRT-PCR in BECs from 10 asthmatic and 10 control individuals. We also sequenced these genes to identify single nucleotide polymorphisms (SNPs), and tested interaction with DNA-me variations. We detected lower DNA-me levels of IL33 and CCL26 in asthmatic than control BECs (Δβ=15%, p<0.001 and Δβ=5%, p=0.036, respectively). Interestingly, carriers of a mutative allele in a haplotype within the promoter of IL33 (formed by chr9:6210100, rs928413 and rs7848215) had a lower DNA-me levels (Δβ=6%, p=0.021 and Δβ=14%, p=0.038 for CpG 1 and mean CpGs 2-3-4, respectively), suggesting a possible interaction between these SNPs and methylation level in IL33 promoter region that may lead to a synergetic risk effect. mRNA levels of the three genes were higher in the asthmatic than non-asthmatic group but did not reach statistical significance. However, an association was observed between CCL26 gene expression and blood eosinophil count in billion per liter (r=0.772; p=0.009). Altogether these findings highlight the importance of investigating both epigenetic and genetic mechanisms (eg. DNA-me, gene expression and polymorphisms) in understanding the epithelial immune response in asthma.
Introduction
Airway epithelium constitutes the first line of host defense against pathogens. Airway epithelial remodeling in asthma involves the disruption of tight junctions, loss of epithelial integrity, impairment of barrier function, goblet cell hyperplasia and metaplasia and mucus hypersecretion; features which correlate with asthma severity.106,107 It is now recognized that epithelial cells are the first set of immune cells interacting with antigens such as aeroallergens in allergic individuals.108 The allergen-epithelial cell interactions release a plethora of inflammatory mediators activating both the innate and adaptive responses; hence, the epithelium plays a key role in the development and persistence of inflammatory response.109
Cytokines derived from the epithelium (e.g. thymic stromal lymphopoietin (TSLP) and interleukins (ILs) -25 and -33)110 have been implicated in asthma pathogenesis. IL-33, a member of the IL-1 family, acts as an endogenous alarm signal to orchestrate the immune response following an epithelium damage.43 In atopic asthma, allergens stimulate the airway epithelium release of IL-33,78 possibly in a severity-dependent manner.111 This leads to an upregulation of TH2-driven inflammation and an activation of the IL-33-specific receptor (IL-1 receptor-like 1 (IL-1RL1)/ST2).109 The TH2 response results in blood eosinophil recruitment, which is highly involved in asthma exacerbations through the liberation of inflammation mediators (e.g. major basic proteins and leukotrienes).80,112,113 Furthermore, IL-33 expression is increased in bronchial epithelial cells (BECs) from asthmatic individuals.78 Additionally, several single nucleotide polymorphisms (SNPs) near and within IL33 and IL1RL1 have been found to associate with asthma and have been validated in several genome-wide association studies (GWASs).2,103-105 IL-33 is known to enhance the production of chemokine (C-C motif) ligand 26 (eotaxin-3; CCL26),82 a potent chemotactic factor for eosinophils.84,85 We previously reported that CCL26 levels are correlated with eosinophil levels in sputum of asthmatic patients and that this relationship is correlated with severity.85 In addition, genetic variants within CCL26 have been found to associate with immunoglobulin (Ig)-E levels.114 Taken together, IL-33 and its downstream target molecules (i.e. IL-1RL1 and CCL26) present themselves as strong candidates for asthma.Epigenetic modifications are suspected to be one type of underlying mechanisms involved in the fine regulation of asthma key gene expression.115 DNA methylation (DNA-me), the most investigated epigenetic modification, is the covalent binding of a methyl group on the fifth carbon of a cytosine pyrimidine ring in response to environmental cues.116 It is most likely to be found when cytosine precedes a guanine (dinucleotide CpG). In addition, several studies highlighted the contribution of DNA-me in processes relevant to asthma physiopathology, including cell differentiation and cytokine production.117-123 In blood cells, DNA-me is involved in the regulation of transcriptional activity of some TH2 genes.118,123-125 However, only few studies assessed whether DNA-me might be altered in the airway epithelium of asthmatic patients126-131 and only one study investigated the interactions between genetic and epigenetic modifications.131 Since the airway epithelium acts as a barrier between the bronchial lumen and lung tissue, and that epigenetic changes are sensitive to environmental insults, DNA-me may be involved in the immune response of asthmatic airway epithelium by modulating the release of cytokines.In this study, we assessed whether DNA-me in the epithelium is involved in the gene expression of IL33, IL1RL1 and CCL26 in asthmatic individuals and looked for evidence of an interplay between epigenetic and genetic mechanisms in gene expression regulation. A better understanding of these mechanisms will bring new insights to new therapeutic development for asthma.
Results
Patient’s characteristics
A total of 20 primary BEC cell lines were used for the study of which 10 were from asthmatic individuals and 10 from healthy subjects.85 All subjects were never-smokers. Of the 10 asthmatic individuals, 6 had mild asthma and 4 had severe eosinophilic asthma (Table 2). The mean age of the severe asthmatic group is significantly higher than the control and mild asthmatic groups. The mean force expiratory volume in 1 second (FEV1) % predicted is 65% (sd=9) for severe asthmatic subjects, 93% (sd=7) for mild subjects and 101% (sd=13) for control subjects. The groups did not differ by blood cell counts.
Epigenotyped loci and surrounding genomic regions
We epigenotyped a total of 14 CpG dinucleotide sites within or upstream (located in promoter region) of IL33, IL1RL1 and CCL26 in DNA isolated from BECs. These loci are shown in Figure 1. IL33 CpG sites 2, 3 and 4, IL1RL1 CpG sites 5 and 6 and CCL26 sites 2 and 3 were correlated (r>0.5; p<0.05). Three of the 14 CpG sites were considered hypomethylated (i.e. <10%; IL1RL1 CpG sites 3, 6 and 7) and were not further analysed. Potential binding sites were detected for several transcription factors (TFs) including GATA binding protein-1 (GATA1), GATA binding protein-3 (GATA3), CCCTC-binfing factor (CTCF), Caudal type homeobox (CDX) and NK2 homeobox 5 (NKX25) (Figure 1).
Epivariations in IL33 and CCL26 are associated with asthma and related phenotypes
We investigated the relationship between DNA-me levels in IL33, IL1RL1 and CCL26 and asthma history with age and sex as covariates in BECs. The promoter region of IL33 (CpG sites 2, 3 and 4) was less methylated in BECs from asthmatic than from non-asthmatic individuals (difference of methylation levels (Δβ) =15%, p<0.001 with Cohen’s effect size (d) =1.1; Figure 2). DNA-me levels at the CCL26 gene promoter region (CpG site 4) were associated with asthma (Δβ=5%, p=0.036 with d=1.3; Figure 2). None of the analyzed CpG sites within IL1RL1 was found to be differentially methylated between asthmatic and non-asthmatic individuals (data not shown).
We next assessed whether DNA-me variations were associated with asthma severity. DNA-me levels of IL33 CpG sites 2-3-4 and CCL26 CpG site 4 were similar between mild asthmatic and eosinophilic severe asthmatic groups (p=0.997 and p=0.053, Supplementary Figure 1A). DNA-me levels were significantly lowered in the severe asthma group compared to the control individuals for the CCL26 CpG site 4 (p=0.002). No association was found between DNA-me levels and the clinical phenotypes FEV1 and cell counts (data not shown).
Gene expression levels of CCL26 is associated with eosinophil count
We investigated the association between IL33, IL1RL1 and CCL26 gene expression in BECs and asthma phenotypes. mRNA levels of IL33, IL1RL1 and CCL26 were higher in the asthmatic than non-asthmatic group but did not reach statistical significance (IL33 fold-change = 4.3, p=0.537; IL1RL1 fold-change = 1.5, p=0.066 and CCL26 fold-change = 1.5, p=0.273, Figure 3A). An association was observed between CCL26 gene expression and blood eosinophil count in billion per liter (r=0.772; p=0.009, Figure 3B); no association was observed for either IL33 or IL1RL1 (data not shown, p=0.683 and p=0.531 respectively). No association was found between gene expression and asthma severity (p=0.992, p=0.356 and p=0.215 for IL33, IL1RL1 and CCL26 respectively, Supplementary Figure 1B).
We assessed the functional impacts of epivariations on transcriptional activity in BECs. No significant correlation was observed between IL33, IL1RL1 and CCL26 DNA-me and mRNA levels (for IL33 CpG 1 and 2-3-4: p=0.390 and p=0.271, for IL1RL1 CpG 1, 2, 4 and 5: p=0.085, 0.106, 0.064 and 0.217, and for CCL26 CpG 1, 2-3 and 4: p=0.295, p=0.393 and p=0.395).
Asthma-associated SNPs are associated with IL33 DNA-me levels
We sequenced the promoter regions of IL33 and CCL26 given the associations detected between DNA-me levels and asthma in these genes (Figure 1 and Table 1). A total of 10 SNPs in IL33 and 2 in CCL26 were identified; of the ten IL33 SNPs 2 were novel (chr9:6210100 and chr9:6215082). Three SNPs in IL33 have too low heterozygote frequency to be analyzed (only 1 heterozygote). Seven SNPs in IL33 were tested for association with asthma and none was found to be associated (p>0.05, Table 1) in the 20 BEC samples. However, association analyses were also performed in the SLSJ cohort using MFQLS analyses and 4 SNPs were associated with asthma including the 2 SNPs associated in the literature (from p=0.020 to p=0.039, Table 1). No association was observed between these SNPs and gene expression of IL33 (Supplementary Table 1). CCL26 SNPs were not tested due to low sample number.
We assessed the associations between the 7 IL33 SNPs and DNA-me levels at IL33 CpG site 1 and IL33 CpG sites 2-3-4 and found that DNA-me levels at CpG site 1 was associated with 5 SNPs (rs992969, chr9:6210100, rs928412, rs928413 and rs7848215) and that DNA-me levels at CpG sites 2-3-4 were associated with 3 SNPs (chr9:6210100, rs928413 and rs7848215) (Table 1). All 7 SNPs were in strong linkage disequilibrium (D’>0.92) (Supplementary Figure 2). IL33 promoter DNA-me levels at CpG site 1 and CpG sites 2-3-4 were 6% and 14% higher in homozygous individuals (homozygous for all SNPs of the haplotypes) compared to heterozygotes (heterozygote for all SNPs of the haplotype), respectively (p=0.021 and 0.038 and d=1.1 and d=1.0; respectively) (Figure 4).
Discussion
In recent years, efforts to better define asthma aetiology have led to the discovery that epigenetic mechanisms modify disease pathogenesis. Here we show that the immune role of airway epithelium is regulated, at least partially, by DNA-me modifications at specific loci within TH2 genes. We and others have previously identified several epivariations within TH2 pro-inflammatory genes in various tissues such as whole blood, CD4(+) and CD8(+) T lymphocytes and umbilical cord DNA.123-125,132,133 Given that the epithelium is significantly affected in asthma,134 an investigation of the epigenetic regulation in BECs is warranted to bring more promising findings in understanding asthma pathogenesis.
Levels
To the best of our knowledge, we are the first to report epivariations in the promoter region of IL33 to be associated with asthma using BECs. These DNA-me levels are likely to be associated with gene expression, suggesting that the epigenotyped CpG sites might be involved in the modulation of IL33 cytokine production by the airway epithelium. Generally speaking, lower DNA-me levels in the promoter induce an upregulation of transcriptional activity;93 however, even though an increase in mRNA levels has been found for the three genes in asthmatic individuals compared to controls, the reverse correlation between DNA-me and mRNA levels did not reach significance for IL33 and CCL26. This may be explained by the great variability observed in asthmatic individuals in this study, particularly for the IL33 gene, and by the small sample size. Given the key role of IL-33 in asthma6,70 and eosinophil-induced airway inflammation135,136 and our findings of lowered DNA-me levels in asthmatic BECs, we expected but did not find a negative correlation between IL33 DNA-me levels and blood eosinophil. The poor correlation between blood and epithelial eosinophil counts may be a contributing factor.137,138 Our finding of hypomethylation in the promoter region of IL33 in asthmatic BECs underscores the need to better define the transcriptional and functional impacts of IL33 epivariations in airway epithelium and the origin of these epigenetic modifications (e.g. foetal development, childhood exposition).109 IL-33 is a pleiotropic alarmin, which is released after stress and mechanical damage;139 hence tissue-specific blockade of IL-33 in asthmatic individuals through DNA-me might be a biologically relevant and efficient therapeutic tool.
We also reported that IL1RL1 is expressed in human BECs. IL-1RL1 is expressed on several target cells such as TH2 lymphocytes, mast cells, type 2 innate lymphoid cells (ILC2s) and eosinophils,140 and its expression on airway epithelial cells has only been reported in mice.141 Although we did not identify an asthma-associated DNA-me signature for IL1RL1, the potential implication of epigenetic modifications in the regulation of epithelial IL1RL1 expression cannot be ruled out as not all the CpG dinucleotide sites within the gene had been epigenotyped. It may be of particular interest to measure other CpG sites as the gene has many alternative splicing sites including a second promoter region for the soluble form, which has an inhibitory function on IL33.142 Furthermore, other DNA regions upstream of the IL1RL1 promoter are likely to regulate transcriptional activities since an enhancer has been identified within this locus (i.e. chr2:102335866-102336151, GRCh38, ENCODE data) and may associate with DNA-me To better assess the presence of a retro-activation or retro-inhibition loop involving IL33 and IL1RL1, it should be interesting to document IL1RL1 expression by other cell types as tissue-infiltrating immune cells (TH2 lymphocytes, mast cells, etc.)140 and to differentiate between the expression of its membrane-bound and soluble forms.142 Epithelial regulation of IL1RL1 release and retro-action on IL33 is crucial to better define, notably because several genetic variants within these genes have been previously associated to asthma pathogenesis and exacerbation, underscoring their pivotal roles.2,105,144,145 Together, these observations highlight the need to better understand the epigenetic regulation of IL1RL1 and how these changes may increase the TH2 inflammation in asthma.
We also reported asthma-associated epivariations within the promoter of CCL26. We showed that DNA-me levels were lower in asthmatic BECs. The roles of CCL26 in the physiopathology of asthma have been widely studied; including our previous report that CCL26 gene expression was negatively correlated with pulmonary function and positively with asthma severity and sputum eosinophil counts.80,83,85 CCL26 also stimulates eosinophil reactive oxygen species and cationic protein releases, both are associated with severe airway epithelium damages in asthmatic patients.80 It is known that CCL26 secretion is mediated by pro-inflammatory cytokines (e.g. IL-4, IL-13 and IL-33) and the TF signal transducer and activator of transcription 6 (STAT6);80-82 however, less is known about the regulation of its gene expression. In patients with eosinophilic esophagitis, another allergic inflammatory disease, lower CCL26 DNA-me and higher gene expression levels have been found in patients’ epithelial cells, suggesting that epigenetic modifications regulate transcriptional activities.146 Nevertheless, further work will be needed to determine if DNA-me is one of the underlying mechanisms involved in the regulation of CCL26 release by the airway epithelium.
Abnormal gene expression of key players (e.g. IL33 and IL1RL1) involved in the TH2 inflammatory response have been found to associate with asthma pathogenesis and exacerbation,147 and our finding of an association between a haplotype within IL33 and DNA-me levels at asthma-associated epivariations (CpG site 1 and CpG sites 2-3-4) within the promoter region suggests an interplay between epigenetic and genetic modifications in asthmatic airways. These SNPs were not associated with asthma in BEC samples. However, the analysis in a larger French-Canadian cohort (the SLSJ cohort) shown an association for two of the three SNPs (the third one was filtered out during quality check). Whether a SNP far from the epivariations could have a biological impact is unclear.148,149 We speculate that a haplotype (formed by chr9:6210100, rs928413 and rs7848215 variants) may interact with DNA-me at the promoter of IL33 and together modulate the asthma risk (i.e. risk effect of the mutative allele). DNA-me mediates gene expression mainly through blocking the access of TFs to their binding sites150,151 and we identified several binding motifs for proteins such as GATA1, GATA3 and CTCF, which have been found to associate with immune response and/or asthma. For example, GATA1 and GATA3 are involved in the promotion of TH2 cytokines expression whereas CTCF mediates the innate immune response in murine models.152-154 These results highlight the probable interactions between genetic and epigenetic modifications and that they synergistically modulate asthma pathogenesis.
The use of primary bronchial cells in this study is biologically relevant given that epithelial damages in the airways are a characteristic feature of asthma. DNA-me and gene expression could be stable among several tissues or can be tissue-specific or cell-type-specific.86 This particularity motivate the use of tissues or cell types directly involved in the pathophysiology studied instead of whole blood samples for example. However, the invasive sampling method and the fastidious laboratory work to obtain BECs limited our sample size and reduced the statistical power of the study. However, in terms of the effect size;155 all significant results had a large Cohen’s effect size (d>0.8); suggesting a biological relevance of our findings. When we investigated the TF binding sites motifs near the asthma-associated epivariations, we identified a motif for CTCF, an insulator generally associated to alternative splicing.156 Hence, further work will be needed to better understand the mechanisms underlying the interaction between DNA-me and TF binding. In addition, we used quantitative pyrosequencing validated for its robustness and reproducibility with documented correlation between replicate of r=0.996 and between laboratories of r=0.98.157
In summary, our findings suggest that IL33 and its downstream target molecule CCL26 modulate asthma risk through DNA-me and their subsequent gene expression. There is evidence of interplay between genetic variants and epigenetic modifications which impact on the asthma risk through a fine-tuning of gene expression. These results underline the need to combine epigenetic and genetic analyses to better understand asthma pathogenesis, especially its severity and the role of airway epithelium.
|
Table des matières
1- Introduction
1.1. Asthme
1.1.1. Description et symptômes
1.1.2. Inflammation
1.1.3. Remodelage des voies respiratoires
1.1.4. Hyperactivité bronchique
1.1.5. Phénotypes
1.1.6. Thérapies actuelles
1.2. Trait complexe
1.2.1. Facteurs génétiques
1.2.2. Épigénétique
1.2.3. Méthylation de l’ADN
1.2.4. Facteurs environnementaux
1.4. Hypothèses et objectifs
2- Chapitre 1 : Methylation profiles of IL33 and CCL26 in bronchial epithelial cells are associated with asthma
2.1. Avant-propos
2.2. Résumé
2.3. Abstract
2.4. Introduction
2.5. Results
2.6. Discussion
2.7. Patients and methods
BEC samples
DNA methylation quantification in BECs
Expression study
Genetic analyses
Identification of potential binding motifs
Statistical analyses
2.8. References
3 – Chapitre 2 – Discussion générale
3.1. Modèles expérimentaux
3.2. Types d’approche expérimentale
3.3. Association épigénétique entre l’interleukine-33 et l’éotaxine-3 et l’asthme
3.4. Associations génétiques et épigénétiques du gène Interleukine 33
3.5. Limites et avantages de l’étude
3.6. Perspectives
4 – Conclusion
Références